The motif viewer program
The motif viewer lets you view all the UTRs associated with yeast genes. It is old (2001) and *very* unpolished,
but maybe gives some idea of what is possible.
To download
Here is a zip file containing all the associated files:
motif_files.zip (2.5M)
Unzip this, and double-click on "motif-viewer.jar" to run. There may be a pause after you double-click and before you
see anything. When it loads, you should see something like this:
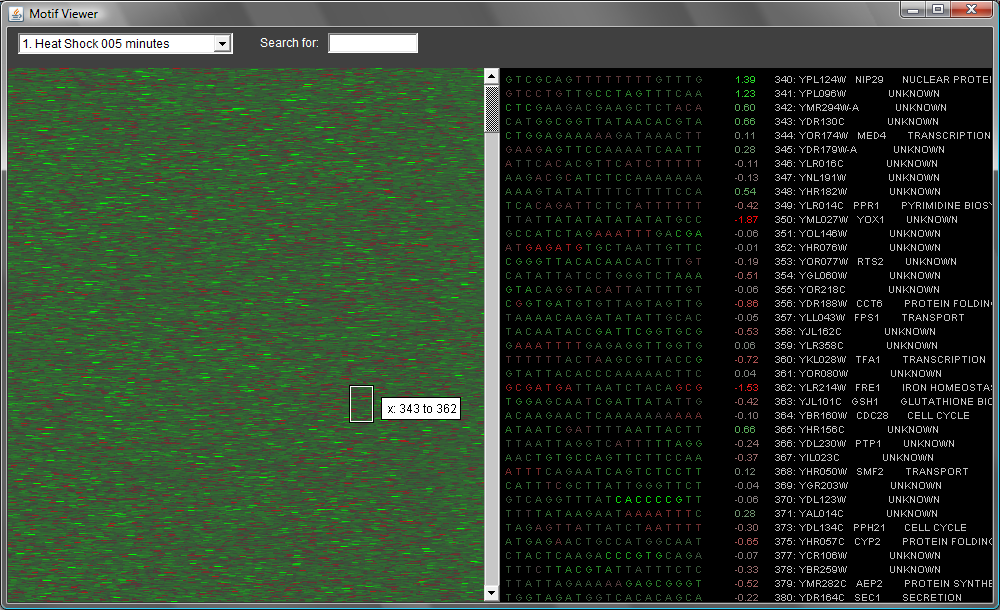
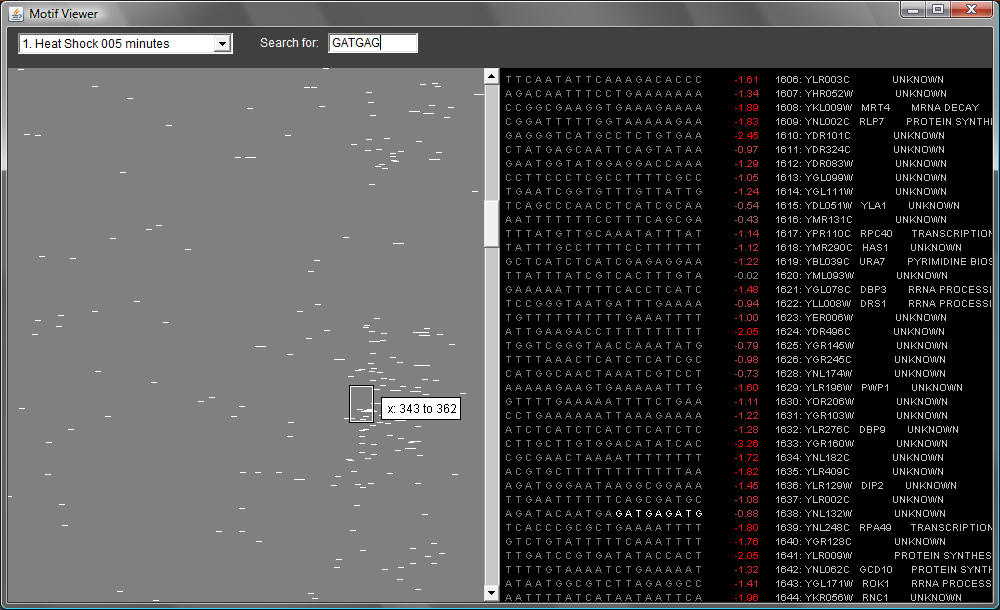
The panel at left is showing all 500-bp upstream transcription regions (UTRs) for yeast genes. Each row represents a different UTR sequence. There is a little rectangle you can drag around; this controls the "detail" view at right. The sequences are colored in such a way that motifs that are often associated with highly expressed genes are green; motifs associated with less expressed genes are red. You can flip through different experimental conditions using the menu at upper left.
You can scroll through all 6,000 genes using the vertical scrollbar in the middle of the screen. If you scroll down, you can see interesting details, as in the image below. (Note that the ordering of the genes is taken from one of Eisen's clusterings, so similarly expressed genes are near each other.)
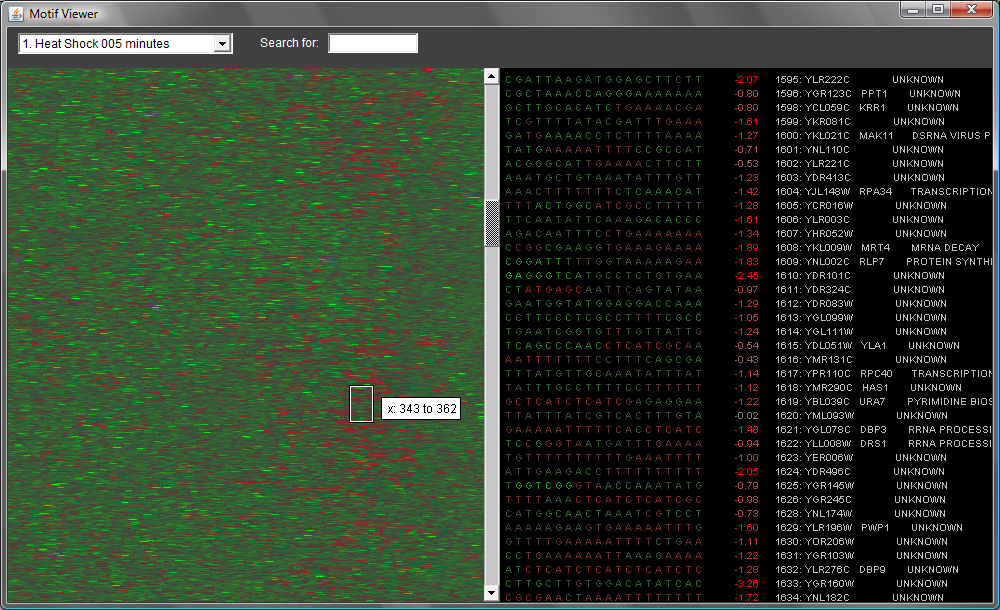
You also can search for a particular motif--type it in the search box at top and hit return. When you do this, you'll see something like the image below. To clear the search, clear the search box and hit return.